The procedures in this section simplify the identification of all 17 basic modifier mechanism. Before using the following operations, which consist merely in clicking either images or descriptive text, it is necessary that the user has determined the dependence on modifier concentration of kcat, Km and kcat/Km.
The procedure to estimate the apparent parameters from initial rate measurements is described in Raw data. Please note that a judicious experimental design with at least 5 substrate and 5 modifier concentrations is indispensable to obtain reliable results for any mechanism, which is not known in advance.
Rigorous kinetic analysis requires determination of the modification mechanism before (!) calculating kinetic parameters by nonlinear regression fitting. Knowing the mechanism, one/two parameters can be constrained to avoid problems associated with parameter intertwining.
The present validation can be run for checking the completeness and internal consistency of kinetic data before submitting a manuscript for publication.
The validity of published data can also be checked if the dependencies of kcat (or limiting rate) and Km can be deduced from graphical and/or tabulated results.
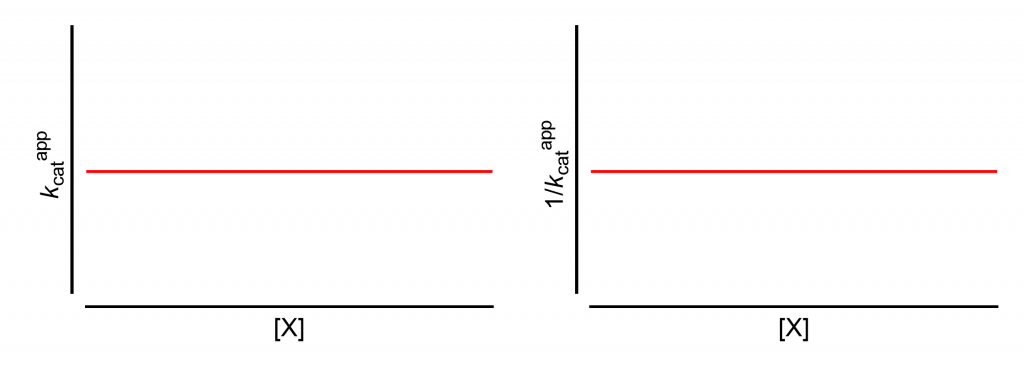
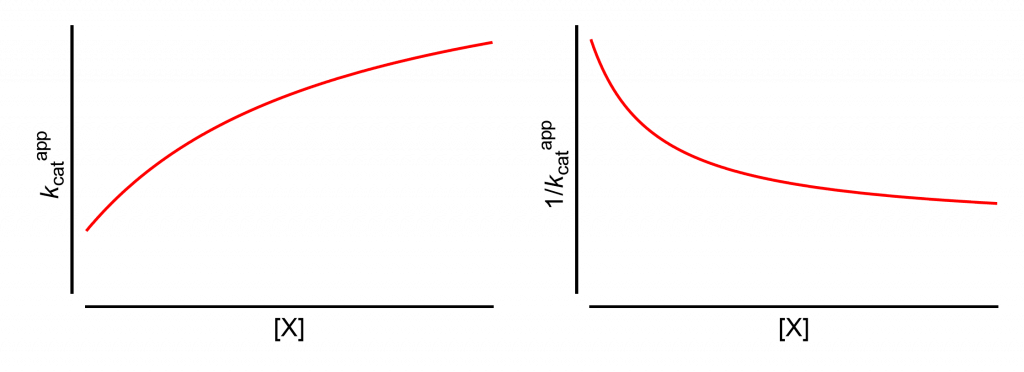
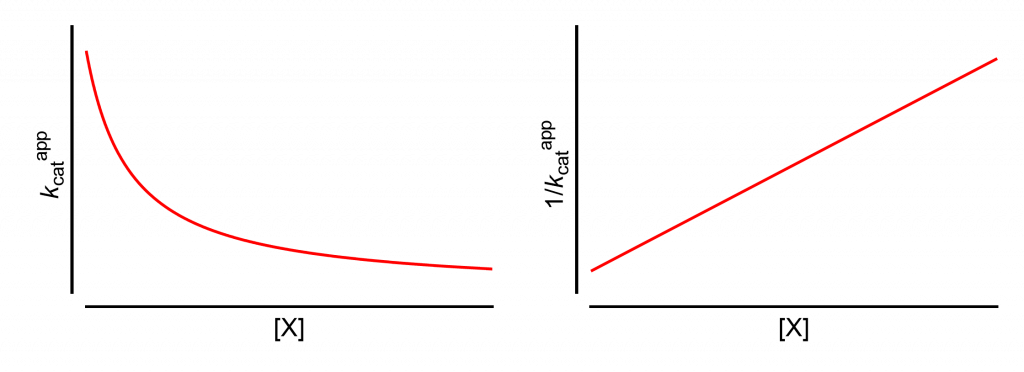
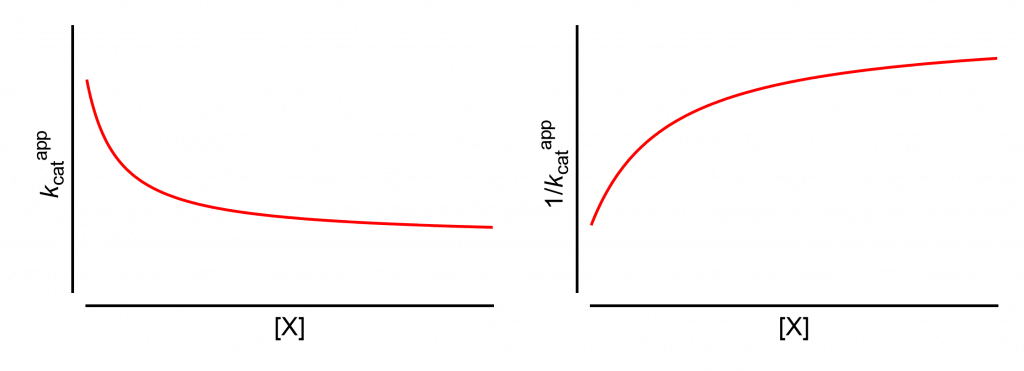
All analyses will start from this page, which shows how the apparent values of kcat and its reciprocal change with increasing modifier concentration, [X].
The same results are obtained if the limiting rate V is used in place of kcat. This is the case when the titrated enzyme active-site concentration is unknown and kcat cannot be calculated.