Hyperbolic specific activation
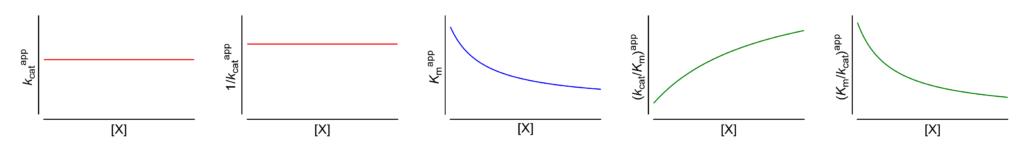
Featured examples
Km↓ The apparent Michaelis constant decreases with increasing [X]
This symbol is shown only when the featured dependence of the parameter on modifier concentration has been demonstrated by the authors.
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors (2) | Reference(3) |
|---|---|---|---|---|---|---|
| 1 | Sucrose α-glucosidase (4) Mus musculus (Balb c) | 3.2.1.48 | Na+ (pH = 7.2) | Sucrose | Pure K-type system, Km↓ α = 0.76, β = 1, KX = 8.79 mM | Gupta (2009) |
| 2 | Sucrose α-glucosidase Rattus norvegicus | 3.2.1.48 | Na+ (pH = 5.4) | Sucrose | K-type activation, Km↓ (5) α = 0.02, β = 1, KX = 39 mM | Semenza (1969) |
| 3 | Sucrose α-glucosidase Hamster (species not specified) | 3.2.1.48 | Na+ (pH = 5.4) | Sucrose | K-type activation, Km↓ (5) α = 0.06, β = 1, KX = 13 mM | Semenza (1969) |
| 4 | 5'-Nucleotidase Legionella pneumophila | 3.1.3.5 | GTP | GMP | K-type activation, Km↓ α = 0.7, β = 1 | Srinivasan (2014) |
| 5 | 1-Phosphatidylinositol phosphodiesterase Bacillus cereus | 3.1.4.10 | Dihexanoylphosphatidylcholine and Butyl-fluorescein myo-inositol phosphate | Butyl-fluorescein myo-inositol phosphate (6) | Activation α' = 0.2, β' = 1.4, Ks = 0.5 mM, K's = 0.15 mM α = 0.03, β = 1.0, KA = 0.2 mM | Birrell (2003) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) The mechanism is species- and pH-dependent, See also under HMx(Sp=Ca)I and HMx(Sp=Ca)A.
(5) Parameters calculated from the data in Fig. 3 (Semenza, 1969).
(6) Symbols as in the original paper [Birrell (2003), free full text, see References]. The enzyme is a small monomeric molecule with a catalytic site and an allosteric site that can bind either a second molecule of substrate, which triggers nonessential activation with mechanism hyperbolic mixed, predominantly catalytic activation [see entry 11 in HMx(Sp<Ca)A], or the activator dihexanoylphosphatidylcholine with mechanism HSpA. The parameters in this page refer to a reaction in the presence of both reactants. Clicking the reference Birrell (2003) below allows to visualize the entire mechanism, made up by reactions schemes A and B acting together.
References
- Birrell GB, Zaikova TO, Rukavishnikov AV, Keana JFW, Hayes Griffith O (2003) Allosteric interactions within subsites of a monomeric enzyme: Kinetics of fluorogenic substrates of PI-specific phospholipase C. Biophys J 84: 3264-3275. doi:10.1016/S0006-3495(03)70051-4
- Gupta S, Mahmood S, Mahmood A (2009) Kinetic characteristics of brush border sucrase activation by Na+ ions in mice intestine. Indian J Exp Biol 47: 811-815.
- Semenza G (1969) A kinetic investigation on the allosteric effects in intestinal sucrase. Eur J Biochem 8: 518-529. doi:10.1111/j.1432-1033.1969.tb00557.x,
- Srinivasan B, Forouhar F, Shukla A, Sampangi C, Kulkarni S, Abashidze M, Seetharaman J, Lew S, Mao L, Acton TB, Xiao R, Everett JK, Montelione GT, Tong L, Balaram H (2014) Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila. FEBS J 281: 1613-1628. doi:10.1111/febs.12727