Linear mixed, predominantly specific inhibition
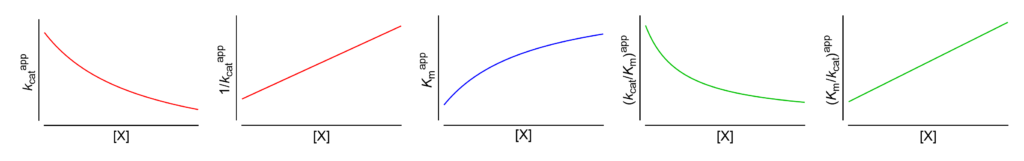
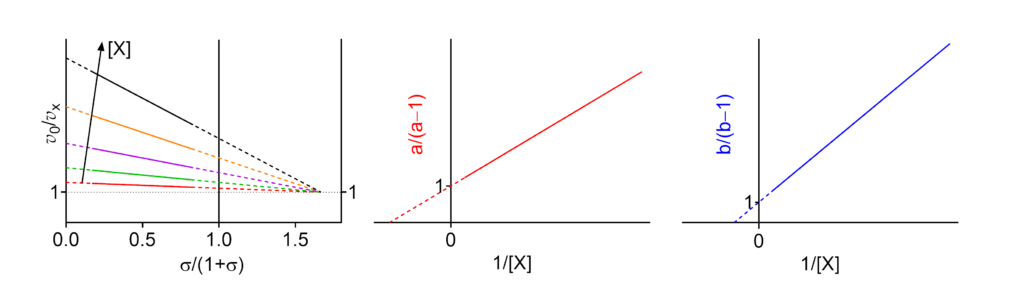
Featured examples
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors(2) | Reference (3) |
|---|---|---|---|---|---|---|
| 1 | Peroxiredoxin Homo sapiens | 1.11.1.15 | Catechol | H2O2 | Partial mixed type noncompetitive inhibition α = 2.2, KX = 1.8 mM | Chow (2016) |
| 2 | Alpha,alpha trehalase Chironomus riparius | 3.2.1.28 | Compound 7 | Trehalose | Linear mixed type inhibition α = 5.8, KX = 34 mM | D'Adamio (2015) |
| 3 | Aminopeptidase N Homo sapiens | 3.4.11.2 | Cholic acid | Leu-Met | Competitive-noncompetitive linear mixed type inhibition α = 9.0, KX = 0.91 mM | Nakanishi (1989) |
| 4 | Aminopeptidase N Homo sapiens | 3.4.11.2 | Deoxycholic acid | Leu-Met | Competitive-noncompetitive linear mixed type inhibition α = 21, KX = 0.42 mM | Nakanishi (1989) |
| 5 | Aminopeptidase N Homo sapiens | 3.4.11.2 | Chenodeoxycholic acid | Leu-Met | Competitive-noncompetitive linear mixed type inhibition α = 7.0, KX = 0.24 mM | Nakanishi (1989) |
| 6 | Aminopeptidase N Homo sapiens | 3.4.11.2 | Deoxycholic acid | Leu-Gly | Competitive-noncompetitive linear mixed type inhibition α = 11, KX = 0.51 mM | Nakanishi (1989) |
| 7 | Acetylcholinesterase Electrophorus electricus | 3.1.1.7 | Pararosaniline | Acetylthiocholine | Linear mixed inhibition α = 3.9, KX = 3.0 μM | Küçükkilinç (2007) |
| 8 | Cholinesterase Homo sapiens | 3.1.1.8 | Pararosaniline | Butyrylthiocholine | Linear mixed inhibition α = 13, KX = 1.9 μM | Küçükkilinç (2005) |
| 9 | Cholinesterase Homo sapiens | 3.1.1.8 | Malachite green | Butyrylthiocholine | Linear mixed inhibition α = 23, KX = 0.28 μM | Küçükkilinç (2005) |
| 10 | Cholinesterase Homo sapiens | 3.1.1.8 | Ethopropazine | Butyrylthiocholine | Linear mixed inhibition α = 8.4, KX = 0.037 μM | Küçükkilinç (2007) |
| 11 | Acetylcholinesterase Electrophorus electricus | 3.1.1.7 | Malachite green | Acetylthiocholine | Linear mixed inhibition α = 4.8, KX = 1.9 μM | Küçükkilinç (2008) |
| 12 | Coagulation factor XIa Homo sapiens | 3.4.21.27 | Aprotinin | Coagulation factor IX | Non-competitive inhibition α = 6.3, KX = 0.89 μM | Ogawa (2005) |
| 13 | Memapsin 2 (4) Homo sapiens | 3.4.23.46 | Pepstatin A methylester pH = 6.5, 22°C | C100 (5) | Linear non-competitive inhibition (6) α = 2.1±1.8, KX = 0.15 μM | Tian (2002) |
| 14 | Memapsin 2 (4) Homo sapiens | 3.4.23.46 | Pepstatin A methylester pH = 6.5, 37°C | C100 (5) | Linear non-competitive inhibition (6) α = 2.0±0.5, KX = 0.71 μM | Tian (2002) |
| 15 | Aldehyde oxidase Homo sapiens | 1.2.3.1 | Domperidone | Phtalazine | Mixed, predominantly competitive inhibition α = 2.6, KX = 5.3 μM | Barr (2011) |
| 16 | Aldehyde oxidase Homo sapiens | 1.2.3.1 | Chlorpromazine | Phtalazine | Mixed, predominantly competitive inhibition α = 5.3, KX = 0.62 μM | Barr (2011) |
| 17 | Aldehyde oxidase Homo sapiens | 1.2.3.1 | Clozapine | Phtalazine | Mixed, predominantly competitive inhibition α = 15, KX = 3.9 μM | Barr (2011) |
| 18 | Spermine synthase Bos taurus | 2.5.1.22 | Spermine (reaction product) | Spermidine | Mixed, predominantly competitive inhibition α = 4, KX = 10 μM | Pajula (1983) (7) |
| 19 | Spermine synthase Bos taurus | 2.5.1.22 | 5'-methylthioadenosine (reaction product) | Decarboxylated adenosylmethionine | Mixed, predominantly competitive inhibition α = 10, KX = 0.2 μM | Pajula (1983) (7) |
| 20 | Tyrosinase (8) Homo sapiens | 1.14.18.1 | Kojic acid | L-Dopa | Mixed, predominantly competitive inhibition α = 3.9, KX = 145 μM | Sun (2014) (9) |
| 21 | Butane-utilizing bacterial cultures (10) | — | Butane | 1,1-dichloroethylene | Mixed inhibition α = 20, KX = 0.23 μM | Kim (2002) |
| 22 | Butane-utilizing bacterial cultures (10) | — | Butane | 1,1-dichloroethane | Mixed inhibition α = 1.8, KX = 1.8 μM | Kim (2002) |
| 23 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Lys2-Tat(1-9) MKPVDPNIE | Ala-Pro-p-nitroanilide | Linear mixed type inhibition α = 10, KX = 42.7 μM | Lorey (2003) (11) |
| 24 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Trp2-Tat(1–9) MWPVDPNIE | Ala-Pro-p-nitroanilide | Linear mixed type inhibition α = 16, KX = 2.12 μM | Lorey (2003) (11) |
| 25 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Met-Trp1-G-CSF(1–8) MWPLGPASS | Ala-Pro-p-nitroanilide | Linear mixed type inhibition α = 16, KX = 12.4 μM | Lorey (2003) (11) |
| 26 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Met-IL-2(1–12) MAPTSSSTKKTQL | Ala-Pro-p-nitroanilide | Linear mixed type inhibition α = 9.4, KX = 269 μM | Lorey (2003) (11) |
| 27 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Met-Trp-Val MWV | Ala-Pro-p-nitroanilide | Linear mixed type inhibition α = 15, KX = 200 μM | Lorey (2003) (11) |
| 28 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Tat(1–9) MDPVDPNIE | Ala-Pro-p-nitroanilide | Parabolic mixed-type inhibition α = 8.9, γ = 0.3, δ =6.5 KX = 267 μM | Lorey (2003) (11)(12) |
| 29 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Tat(1–9) MDPVDPNIE | Gly-Pro-R110-CO-(CH2)4Cl (13) | Parabolic mixed-type inhibition α = 0.8, γ = 0.8, δ =2.2 KX = 230 μM | Lorey (2003) (11)(12) |
| 30 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Trp1-Tat(1–9) WDPVDPNIE | Ala-Pro-p-nitroanilide | Parabolic mixed-type inhibition α = 46, γ = 1.5, δ =15 KX = 150 μM | Lorey (2003) (11)(12) |
| 31 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Gly3-Tat(1–9) MDGVDPNIE | Ala-Pro-p-nitroanilide | Parabolic mixed-type inhibition α = 3.7, γ = 0.3, δ =2.2 KX = 487 μM | Lorey (2003) (11)(12) |
| 32 | Dipeptidyl-peptidase IV Homo sapiens | 3.4.14.5 | Ile3-Tat(1–9) MDIVDPNIE | Ala-Pro-p-nitroanilide | Parabolic mixed-type inhibition α = 1.7, γ = 9.2, δ =0.01 KX = 1.75 mM | Lorey (2003) (11)(12) |
| 33 | β-Lactamase Bacillus cereus 5B6 | 3.5.2.6 | Compound 6a | Nitrocefin | Mixed type inhibition α = 3.8, KX = 6.6 μM | Siemann (2002) |
| 34 | β-Lactamase Bacillus cereus 5B6 | 3.5.2.6 | Compound 11c | Nitrocefin | Mixed type inhibition α = 3.1, KX = 0.7 μM | Siemann (2002) |
| 35 | Catechol 1,2-dioxygenase Acinetobacter baylyi | 1.13.11.1 (13) | 4-Chloroaniline | 3,4-Dichloroaniline | Mixed inhibition α = 1.14, KX = 101 μM | Hongsawat (2011) |
| 36 | Cruzipain Tripanosoma cruzi | 3.4.22.51 | Fukugetin | Cbz-Phe-Arg-7-amino-4-methyl coumarylamide | Partial competitive inhibition α = 4.5, KX = 1.0 μM | Assis (2013) |
| 37 | S-(hydroxymethyl)gluta- thione dehydrogenase Homo sapiens | 1.1.1.284 | Resveratrol | Doxorubicin | Mixed type inhibition α = 2.98, KX = 55.8 μM | Ito (2013) |
| 38 | Linoleate 13S-lipoxygenase Glycine max | 1.13.11.12 | (Z)-9-palmitoleyl sulfate | Linoleic acid | Linear mixed inhibition α = 10.2, KX = 13.7 μM | Ruddat (2003) |
| 39 | NADH:ubiquinone reductase (H+-translocating) Escherichia coli | 1.6.5.3 | Zn2+ | NADred | Linear mixed inhibition α = 1.7, KX = 50 μM | Schulte (2014) |
| 40 | Neurolysin Rattus norvegicus | 3.4.24.16 | R2 | Internally quenched peptide containing the neurotensin sequence | Mixed inhibition, noncompetitive and competitive components α = 6.1, KX = 18 μM | Hines (2014) |
| 41 | Adenylate cyclase Homo sapiens | 4.6.1.1 | Bithionol | ATP | Mixed type inhibition α = 1.7, KX = 2.3 μM | Kleinboelting (2016) |
| 42 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Edrophonium | Acetylthiocholine | Linear mixed inhibition α = 7.1, KX = 0.15 μM | Berman (1990) |
| 43 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Phenyltrimethylammonium | Acetylthiocholine | Linear mixed inhibition α = 5.9, KX = 20 μM | Berman (1990) |
| 44 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | N-Methylacridinium | Acetylthiocholine | Linear mixed inhibition α = 1.9, KX = 46 μM | Berman (1990) |
| 45 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Decamethonium | Acetylthiocholine | Linear mixed inhibition α = 3.2, KX = 0.56 μM | Pietsch (2005) |
| 46 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Hexamethonium | Acetylthiocholine | Linear mixed inhibition α = 4.2, KX = 13 μM | Pietsch (2005) |
| 47 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Decyltrimethylammonium | Acetylthiocholine | Linear mixed inhibition α = 8.4, KX = 34 μM | Pietsch (2005) |
| 48 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Hexyltrimethylammonium | Acetylthiocholine | Linear mixed inhibition α = 3.3, KX = 142 μM | Pietsch (2005) |
| 49 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Propidium | Acetylthiocholine | Linear mixed inhibition α = 1.6, KX = 1.1 μM | Pietsch (2005) |
| 50 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | d-Tubocurarine | Acetylthiocholine | Linear mixed inhibition α = 3.4, KX = 40 μM | Pietsch (2005) |
| 51 | Acetylcholinesterase Electrophorus electricus | 3.1.1.7 | Tacrine | Acetylthiocholine | Linear mixed type inhibition α = 1.48, KX = 25 μM | Pietsch (2005) |
| 52 | Acetylcholinesterase Electrophorus electricus | 3.1.1.7 | Galanthamine | Acetylthiocholine | Linear mixed type inhibition α = 2.0, KX = 37 μM | Pietsch (2005) |
| 53 | Protein-glutamine gamma-glutamyl- transferase (14) Homo sapiens | 2.3.2.13 | Compound 5c | Compound 5b | Linear mixed, predominantly specific inhibition α = 2.0, KX = 585 μM | Wodtke (2016) (15) |
| 54 | Tyrosinase Agaricus bisporus | 1.14.18.1 | Kojic acid | L-Dopa | Competitive-noncompetitive inhibition α = 2.0, KX = 37 μM | Liu (2017) |
| 55 | ABC-type quaternary amine transporter Homo sapiens | 7.6.2.9 | Compound 9 | γ-Aminobutyric acid | Competitive inhibition | Al-Khawaja (2014) |
| 56 | Proton-translocating NAD(P)+ transhydrogenase Escherichia coli | 7.1.1.1 | 5'-AMP | NADPred (at fixed 3-acetylpyridine adenine dinucleotide) | Noncompetitive inhibition α = 2.7, KX = 3.7 μM | Hanson (1979) |
| 57 | Pyridoxal 5'-phosphate synthase (wild type) Escherichia coli | 1.4.3.5 | Pyridoxal 5'-phosphate (reaction product) | Pyridoxine 5'-phosphate | Mixed-type inhibition α = 12-14, KX = 7.1 μM | Barile (16) (2019) |
| 58 | beta-Galactosidase Escherichia coli | 3.2.1.23 | Glycofullerene 10 (17) | p-Nitrophenyl β-D-Galactopyranoside | Mixed-type inhibition α = 6.1, KX = 1.8 μM | Abellán Flos (2016) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) Memapsin 2 is the recommended name of an enzyme with 82 synonyms (BRENDA, July 2016), including β-secretase and γ-secretase. It is part of a complex membrane-bound macromolecular complex that involves presenilins 1 and 2.
(5) C100 = Recombinant protein with amino acid sequence identical to the C-terminal fragment of presenilin (cleaved by memapsin 2 from the amyloid precursor protein) bearing an extra methionine residue at the N-terminus.
(6) In this study Tian et al. (2002) paid particular attention to accurate determinations of Kis and Kii, the specific and the catalytic components of the mechanisms, respectively, providing errors and graphical representations. The linearity of inhibition was ascertained experimentally. Taking into account error propagation, these data have been used in this table to calculate α as Kii/Kis with its associated error that, together with graphics, was useful to identify the mechanism. The authors called all mechanisms ‘linear non-competitive inhibition’, which belong however to LMx(Sp>Ca)I, LMx(Sp<Ca)I and LMx(Sp=Ca)I. See also the tables of LMx(Sp<Ca)I and LMx(Sp=Ca)I.
(7) A study dedicated to inhibition of spermine synthase (two substrates-two products reaction) by reaction products that suggests a compulsory-order mechanism in which both substrates bind the enzyme before release of products. See also the pages of LMx(Sp<Ca)I and LSpI.
(8) In this study a truncated, His-tagged form of tyrosinase, which includes the catalytic domain, was used.
(9) This paper by Sun and coworkers is a recommended reading because it is a useful tutorial in the management of enzyme-inhibition kinetic data. The authors fit the general modifier mechanism of Botts and Morales to an entire ensemble of data with a large number of experimental points. The analysis of inhibition mechanism is based on graphical representation of data and on statistical test that include the Akaike information criterion. See also other results from this study under LSpI and LMx(Sp<Ca)I.
(10) Aerobic cometabolism cultures of Gram-positive and Gram-negative bacteria aimed at decontaminating groundwater from chlorinated hydrocarbons. Despite the enzymes responsible for the decontaminating reactions could not be identified, the inhibition results are clear and support the optimization of bacterial growth conditions in plants of water decontamination.
(11) The study by Lorey et al. (2003) is centered on the inhibition of human dipeptidyl-peptidase IV by the peptide MDPVDPNIE, Tat(1-9), corresponding to the N-terminus of the human immunodeficiency virus-1 transactivator Tat (HIV-1 Tat, which consists of 86 amino acids), and derived peptides. See also cases of linear specific inhibition at the page LSpI.
(12) The entries with colored background represent a mechanism, called by the authors parabolic mixed-type inhibition, which has its roots in linear mixed, predominantly specific inhibition, LMx(Sp>Ca)I. The term parabolic refers to the curved-up slope replot of double-reciprocal plots but the inhibitory nature is still linear since the reaction rate approaches zero at saturating inhibitor concentrations. Further details HERE.
(13) And possibly similar activities in the bacterial culture.
(14) Synonyms: Tissue transglutaminase 2, Transglutaminase 2.
(15) This is the first publication in which the systematics of enzyme-modifier interactions proposed by Baici, including the nomenclature, has been adopted.
(16) A significant example of allosteric product inhibition. This paper is a recommended reading for accurate experimental design, data analysis and discussion that strongly support the results. The mechanism was unequivocally assigned on the basis of all dependencies of the apparent kinetic parameters on modifier concentration.
(17) Complex structure: see Abellán Flos et al. (2016), Supplementary Information, p. 19.
References
- Abellán Flos M, García Moreno MI, Ortiz Mellet C, García Fernández JM, Nierengarten J-F, Vincent SP (2016) Potent glycosidase inhibition with heterovalent fullerenes: Unveiling the binding modes triggering multivalent inhibition. Chem Eur J 22: 11450-11460. doi:10.1002/chem.201601673
- Al-Khawaja A, Petersen JG, Damgaard M, Jensen MH, Vogensen SB, Lie MEK, Kragholm B, Bräuner-Osborne H, Clausen RP, Frølund B, Wellendorph P (2014) Pharmacological identification of a guanidine-containing β-alanine analogue with low micromolar potency and selectivity for the betaine/GABA transporter 1 (BGT1). Neurochem Res 39: 1988-1996. doi:10.1007/s11064-014-1336-9
- Assis DM, Gontijo VS, de Oliveira Pereira I, Santos JA, Camps I, Nagem TJ, Ellena J, Izidoro MA, dos Santos Tersariol IL, de Barros NM, Doriguetto AC, dos Santos MH et al. (2013) Inhibition of cysteine proteases by a natural biflavone: behavioral evaluation of fukugetin as papain and cruzain inhibitor. J Enzyme Inhib Med Chem 28: 661-670. doi:10.3109/14756366.2012.668539
- Barile A, Tramonti A, di Salvo ML, Nogués I, Nardella C, Malatesta F, Contestabile R (2019) Allosteric feedback inhibition of pyridoxine 5′-phosphate oxidase from Escherichia coli. J Biol Chem 294: 15593-15603. doi:10.1074/jbc.RA119.009697
- Barr JT, Jones JP (2011) Inhibition of human liver aldehyde oxidase: implications for potential drug-drug interactions. Drug Metab Disp 39: 2381-2386. doi:10.1124/dmd.111.041806
- Berman HA, Leonard K (1990) Ligand exclusion on acetylcholinesterase. Biochemistry 29: 10640-10649. doi:10.1021/bi00499a010
- Chow ML, Troussicot L, Martin M, Doumèche B, Guillière F, Lancelin J-M (2016) Predicting and understanding the enzymatic inhibition of human peroxiredoxin 5 by 4-substituted pyrocatechols by combining funnel metadynamics, solution NMR, and steady-state kinetics. Biochemistry 55: 3469-3480. doi:10.1021/acs.biochem.6b00367
- D’Adamio G, Sgambato A, Forcella M, Caccia S, Parmeggiani C, Casartelli M, Parenti P, Bini D, Cipolla L, Fusi P, Cardona F (2015) New synthesis and biological evaluation of uniflorine A derivatives: towards specific insect trehalase inhibitors. Org Biomol Chem 13: 886-892. doi:10.1039/C4OB02016B
- Hanson RL (1979) The kinetic mechanism of pyridine nucleotide transhydrogenase from Escherichia coli. J Biol Chem 254: 888-893.
- Hines CS, Ray K, Schmidt JJ, Xiong F, Feenstra RW, Pras-Raves M, de Moes JP, Lange JHM, Melikishvili M, Fried MG, Mortenson P, Charlton M et al. (2014) Allosteric Inhibition of the Neuropeptidase Neurolysin. J Biol Chem 289: 35605-35619. doi:10.1074/jbc.M114.620930
- Hongsawat P, Vangnai AS (2011) Biodegradation pathways of chloroanilines by Acinetobacter baylyi strain GFJ2. J Hazard Mater 186: 1300-1307. doi:10.1016/j.jhazmat.2010.12.002
- Ito Y, Mitani T, Harada N, Isayama A, Tanimori S, Takenaka S, Nakano Y, Inui H, Yamaji R (2013) Identification of carbonyl reductase 1 as a resveratrol-binding protein by affinity chromatography using 4′-amino-3,5-dihydroxy-trans-stilbene. Journal of Nutritional Science and Vitaminology (Tokyo) 59: 358-364. doi:10.3177/jnsv.59.358
- Kim Y, Arp DJ, Semprini L (2002) Kinetic and inhibition studies for the aerobic cometabolism of 1,1,1-trichloroethane, 1,1-dichloroethylene, and 1,1-dichloroethane by a butane-grown mixed culture. Biotechnol Bioeng 80: 498-508. doi:10.1002/bit.10397
- Kleinboelting S, Ramos-Espiritu L, Buck H, Colis L, van den Heuvel J, Glickman JF, Levin LR, Buck J, Steegborn C (2016) Bithionol potently inhibits human soluble adenylyl cyclase through binding to the allosteric activator site. J Biol Chem 291: 9776-9784. doi:10.1074/jbc.M115.708255
- Küçükkilinç T, Özer I (2005) Inhibition of human plasma cholinesterase by malachite green and related triarylmethane dyes: mechanistic implications. Arch Biochem Biophys 440: 118-122. doi:10.1016/j.abb.2005.06.003
- Küçükkilinç T, Özer I (2007) Multi-site inhibition of human plasma cholinesterase by cationic phenoxazine and phenothiazine dyes. Arch Biochem Biophys 461: 294-298. doi:10.1016/j.abb.2007.02.029
- Küçükkilinç TT, Ozer I (2008) Inhibition of electric eel acetylcholinesterase by triarylmethane dyes. Chem-Biol Interact 175: 309-311. doi:10.1016/j.cbi.2008.05.008
- Liu D-M, Yang J-L, Ha W, Chen J, Shi Y-P (2017) Kinetics and inhibition study of tyrosinase by pressure mediated microanalysis. Anal Biochem 525: 54-59. doi:10.1016/j.ab.2017.02.020
- Lorey S, Stöckel-Maschek A, Faust J, Brandt W, Stiebitz B, Gorrell MD, Kähne T, Mrestani-Klaus C, Wrenger S, Reinhold D, Ansorge S, Neubert K (2003) Different modes of dipeptidyl peptidase IV (CD26) inhibition by oligopeptides derived from the N-terminus of HIV-1 Tat indicate at least two inhibitor binding sites. Eur J Biochem 270: 2147-2156. doi:10.1046/j.1432-1033.2003.03568.x
- Nakanishi M, Moriyama A, Narita Y, Sasaki M (1989) Aminopeptidase-M from human liver. II. Kinetic analysis of inhibition of the enzyme by bile acids. J Biochem 106: 826-830. doi:10.1093/oxfordjournals.jbchem.a122938
- Ogawa T, Verhamme IM, Sun MF, Bock PE, Gailani D (2005) Exosite-mediated substrate recognition of factor IX by factor XIa. The factor XIa heavy chain is required for initial recognition of factor IX. J Biol Chem 280: 23523-23530. doi:10.1074/jbc.M500894200
- Pajula RL (1983) Kinetic properties of spermine synthase from bovine brain. Biochem J 215: 669-676. doi:10.1042/bj2150669
- Pietsch M, Gütschow M (2005) Synthesis of tricyclic 1,3-oxazin-4-ones and kinetic analysis of cholesterol esterase and acetylcholinesterase inhibition. J Med Chem 48: 8270-8288. doi:10.1021/jm0508639
- Ruddat VC, Whitman S, Holman TR, Bernasconi CF (2003) Stopped-flow kinetic investigations of the activation of soybean lipoxygenase-1 and the influence of inhibitors on the allosteric site. Biochemistry 42: 4172-4178. doi:10.1021/bi020698o
- Schulte M, Mattay D, Kriegel S, Hellwig P, Friedrich T (2014) Inhibition of Escherichia coli respiratory complex I by Zn2+. Biochemistry 53: 6332-6339. doi:10.1021/bi5009276
- Siemann S, Evanoff DP, Marrone L, Clarke AJ, Viswanatha T, Dmitrienko GI (2002) N-arylsulfonyl hydrazones as inhibitors of IMP-1 metallo-beta-lactamase. Antimicrobial agents and chemotherapy 46: 2450-2457. doi:10.1128/AAC.46.8.2450-2457.2002
- Sun W, Wendt M, Klebe G, Röhm KH (2014) On the interpretation of tyrosinase inhibition kinetics. J Enzyme Inhib Med Chem 29: 92-99. doi:10.3109/14756366.2012.755621
- Tian GC, Sobotka-Briner CD, Zysk J, Liu XD, Birr C, Sylvester MA, Edwards PD, Scott CD, Greenberg BD (2002) Linear non-competitive inhibition of solubilized human gamma-secretase by pepstatin a methylester, L685458, sulfonamides, and benzodiazepines. J Biol Chem 277: 31499-31505. doi:10.1074/jbc.M112328200
- Wodtke R, Schramm G, Pietzsch J, Pietsch M, Löser R (2016) Synthesis and kinetic characterisation of water-soluble fluorogenic acyl donors for transglutaminase 2. ChemBioChem 17: 1263-1281. [Corrigendum: ChemBioChem 2016, 17, 1674]. doi:10.1002/cbic.201600048