Hyperbolic catalytic activation
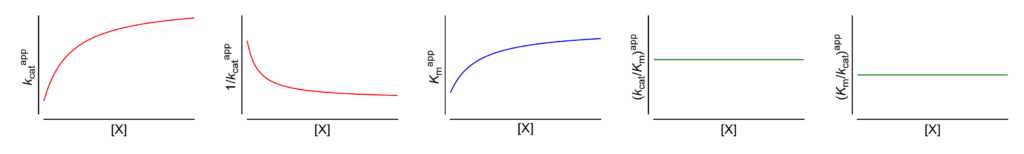
Featured examples
- V/Km ⫫ (∴ kcat/Km ⫫) The apparent specificity constant is independent of [X]
- Km/V ⫫ (∴ Km/kcat ⫫) The apparent reciprocal specificity constant is independent of [X]
- Km↑ The apparent Michaelis constant increases with increasing [X]
- V ↑ (∴ kcat↑) The apparent limiting rate, and therefore the catalytic constant, increase with increasing [X]
These symbols are shown only when the featured dependencies of the parameters on modifier concentration have been demonstrated by the authors.
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors (2) | Reference(3) |
|---|---|---|---|---|---|---|
| 1 | Glutamate dehydrogenase (NAD(P)+) Not specified | 1.4.1.3 | ADP | NADPred | Uncompetitive activating behavior. Clear diagnosis, parameters imprecise. | Frieden (1963) |
| 2 | Thioglucosidase Raphanus sativus | 3.2.1.147 | Ascorbate | Sinigrin | Linear uncompetitive activation (4) Km↑, V↑ | Shikita (1999) |
| 3 | Amidase Pseudomonas aeruginosa | 3.5.1.4 | Hydroxylamine | Acetamide | Uncompetitive activation Km↑, V↑, α = β = 9.2, KX = 16.8 mM | Woods (1979) |
| 4 | Peroxidase Armoracia rusticana | 1.11.1.7 | Melamine | o-Phenylenediamine pH = 7.4 | Noncompetitive activation Km↑, V↑, V/Km ⫫, Km/V ⫫ (5) | Karaseva (2003) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) From the graphics in Figure 4 (Shikita, 1999) the mechanism is clearly nonessential activation, which cannot be linear, and the correct name is hyperbolic catalytic activation. The allosteric coupling constant and the activation constant KX have not been calculated. The confusion about uncompetitive interactions in this and other publications is due to misinterpretation of the basic mechanisms as discussed elsewhere (pp. 150-153 and 273-276)
(5) The last two properties deduced from Fig. 7 in the paper. At pH = 4.3 the mechanism changes from activation to inhibition with mechanism HMx(Sp<Ca)I.
References
- Frieden C (1963) Glutamate dehydrogenase. V. The relation of enzyme structure to the catalytic function. J Biol Chem 238: 3286-3299.
- Karaseva EI, Naumchik IV, Metelitza DI (2003) Noncompetitive activation of the peroxidase-catalyzed oxidation of o-phenylenediamine by melamine (in Russian). Bioorganicheskaya Khimiya 29: 49-56. doi:10.1023/A:1022278402568
- Shikita M, Fahey JW, Golden TR, Holtzclaw WD, Talalay P (1999) An unusual case of ‘uncompetitive activation’ by ascorbic acid: purification and kinetic properties of a myrosinase from Raphanus sativus seedlings. Biochem J 341: 725-732. doi:10.1042/bj3410725
- Woods MJ, Findlater JD, Orsi BA (1979) Kinetic mechanism of the aliphatic amidase from Pseudomonas aeruginosa. Biochim Biophys Acta 567: 225-237. doi:10.1016/0005-2744(79)90189-X