Hyperbolic catalytic inhibition
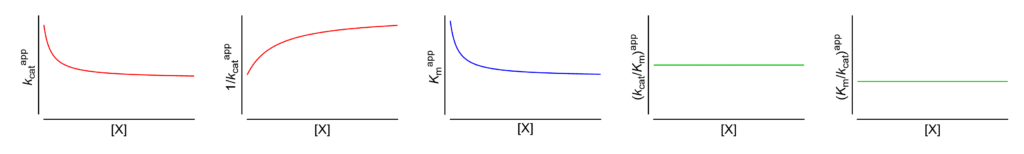
Featured examples
- Km↓ The apparent Michaelis constant decreases with increasing [X]
- V ↓ (∴kcat ↓) The apparent limiting rate, and therefore the catalytic constant, decrease with increasing [X]
- (V/Km)⫫ (∴kcat/Km ⫫) The apparent V/Km ratio, and therefore the specificity constant, are independent of [X]
These symbols are shown only when the featured dependencies of the parameters on modifier concentration have been demonstrated by the authors.
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors (2) | Reference(3) |
|---|---|---|---|---|---|---|
| 1 | Glutamate dehydrogenase (NAD(P)+) Not specified | 1.4.1.3 | GTP | NADPred | Uncompetitive inhibitory behavior. Clear diagnosis, parameters imprecise | Frieden (1962, 1963) |
| 2 | Cathepsin K Homo sapiens | 3.4.22.38 | Compound 7 | Cbz-Phe-Arg-7-amino-4-methylcoumarylamide | Hyperbolic uncompetitive inhibition α = β = 0.19, KX = 0.29 mM | Novinec (2014) |
| 3 | Cathepsin K Homo sapiens | 3.4.22.38 | Compound 8 | Cbz-Phe-Arg-7-amino-4-methylcoumarylamide | Hyperbolic uncompetitive inhibition α = β = 0.31, KX = 1.1 mM | Novinec (2014) |
| 4 | Penicillin amidase Escherichia coli ATC11105 | 3.5.1.11 | PEG-400 (4) | p-dimethylamino-benzaldehyde | Hyperbolic uncompetitive inhibition α = β = 0.28, KX = 142 mM | Kazan (1997) |
| 5 | Penicillin amidase Escherichia coli ATC11105 | 3.5.1.11 | PEG-4000 (4) | p-dimethylamino-benzaldehyde | Hyperbolic uncompetitive inhibition α = β = 0.34, KX = 30.7 mM | Kazan (1997) |
| 6 | Penicillin amidase Escherichia coli ATC11105 | 3.5.1.11 | PEG-10,000 (4) | p-dimethylamino-benzaldehyde | Hyperbolic uncompetitive inhibition α = β = 0.17, KX = 45.8 mM | Kazan (1997) |
| 7 | Glutathione-disulfide reductase Saccharomyces cerevisiae | 1.8.1.7 | Safranin O | L-Glutathione (oxidized) | Hyperbolic uncompetitive inhibition α = β = 0.15, KX = 500 μM | Lüönd (1998) |
| 8 | Glutathione-disulfide reductase Saccharomyces cerevisiae | 1.8.1.7 | Thionin O | L-Glutathione (oxidized) | Hyperbolic uncompetitive inhibition α = β = 0.15, KX = 0.4 μM | Lüönd (1998) |
| 9 | Glutathione-disulfide reductase Saccharomyces cerevisiae | 1.8.1.7 | 6-Anilino-5,8-quinolinedione (5) | L-Glutathione (oxidized) | Hyperbolic uncompetitive inhibition α = β = 0.14, KX = 14 μM | Lüönd (1998) |
| 10 | Glutathione-disulfide reductase Saccharomyces cerevisiae | 1.8.1.7 | 2-Anilino-1,4-naphthoquinone | L-Glutathione (oxidized) | Hyperbolic uncompetitive inhibition α = β = 0.19, KX = 21 μM | Lüönd (1998) |
| 11 | Xanthine oxidase Bos taurus | 1.2.3.2 | Xanthine (substrate) | O2 | Hyperbolic uncompetitive inhibition (6) | Morpeth (1983) |
| 12 | NADH:ubiquinone reductase (non-electrogenic) Saccharomyces cerevisiae | 1.6.5.9 | Flavone | NADred | Hyperbolic uncompetitive inhibition α = β < 1, KX = 5.4 μM | Velázquez (2001) |
| 13 | 4-Hydroxy-tetrahydro-dipicolinate synthase Escherichia coli | 4.3.3.7 | Lysine | Pyruvate | Partial mixed inhibition α = β = 0.046, KX = 3.9 mM | Dobson (2004) |
| 14 | Cathepsin B Homo sapiens | 3.4.22.1 | Cyclopalladated bisphosphinic complex | Cbz-Phe-Arg-7-amino-4-methylcoumarylamide | Hyperbolic mixed type inhibition Km↓, V↓, and α = β = 0.18-0.19 | Bincoletto (2005) |
| 15 | Thrombin Homo sapiens | 3.4.21.5 | Hirudin45-65 fragment | Tos-Gly-Pro-Arg-7-amino-4-methylcoumarylamide | Hyperbolic uncompetitive inhibition α = β = 0.49, KX = 0.71 μM | Schmitz (1991) |
| 16 | Lipopolysaccharide heptosyltransferase I Escherichia coli | 2.4.99.B6 | Hexene-beta-D-glucopyranoside | O-deacylated E. coli Kdo2-Lipid A | Uncompetitive inhibition (7) Km↓, V↓, (V/Km)⫫ | Nkosana (2018) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) PEG-400, PEG-4000, PEG-10,000 = polyethylene glycol with molecular mass.
(5) In the referred paper, the molecule with the LY83583 acronym.
(6) Substrate inhibition.
(7) The measurements were performed with only three modifier concentrations (10, 100 and 1000 μM), which are barely sufficient for diagnosing the mechanism but inadequate for parameter calculations.
References
- Bincoletto C, Tersariol ILS, Oliveira CR, Dreher S, Fausto DM, Soufen MA, Nascimento FD, Caires ACF (2005) Chiral cyclopalladated complexes derived from N,N-dimethyl-1-phenethylamine with bridging bis(diphenylphosphine)ferrocene ligand as inhibitors of the cathepsin B activity and as antitumoral agents. Bioorg Med Chem 13: 3047-3055. doi:10.1016/j.bmc.2005.01.057
- Dobson RCJ, Griffin MDW, Roberts SJ, Gerrard JA (2004) Dihydrodipicolinate synthase (DHDPS) from Escherichia coli displays partial mixed inhibition with respect to its first substrate, pyruvate. Biochimie 86: 311-315. doi:10.1016/j.biochi.2004.03.008
- Frieden C (1962) The unusual inhibition of glutamate dehydrogenase by guanosine di- and triphosphate. Biochim Biophys Acta 59: 484-486. doi:10.1016/0006-3002(62)90204-4
- Frieden C (1963) Glutamate dehydrogenase. V. The relation of enzyme structure to the catalytic function. J Biol Chem 238: 3286-3299.
- Kazan D, Erarslan A (1997) Stabilization of Escherichia coli penicillin G acylase by polyethylene glycols against thermal inactivation. Appl Biochem Biotechnol 62: 1-13. doi:10.1007/BF02787979
- Lüönd RM, McKie JH, Douglas KT, Dascombe MJ, Vale J (1998) Inhibitors of glutathione reductase as potential antimalarial drugs. Kinetic cooperativity and effect of dimethyl sulphoxide on inhibition kinetics. J Enzyme Inhib 13: 327-345. doi:10.3109/14756369809021479
- Morpeth FF (1983) Studies on the specificity toward aldehyde substrates and steady-state kinetics of xanthine oxidase. Biochim Biophys Acta 744: 328-334. doi:10.1016/0167-4838(83)90207-8
- Nkosana NK, Czyzyk DJ, Siegel ZS, Cote JM, Taylor EA (2018) Synthesis, kinetics and inhibition of Escherichia coli Heptosyltransferase I by monosaccharide analogues of Lipid A. Bioorg Med Chem Lett 28: 594-600. doi:10.1016/j.bmcl.2018.01.040
- Novinec M, Lenarčič B, Baici A (2014) Probing the activity modification space of the cysteine peptidase cathepsin K with novel allosteric modifiers. PLoS One 9: e106642. doi:10.1371/journal.pone.0106642
- Schmitz T, Rothe M, Dodt J (1991) Mechanism of the inhibition of a-thrombin by hirudin-derived fragments hirudin(1-47) and hirudin(45-65). Eur J Biochem 195: 251-256. doi:10.1111/j.1432-1033.1991.tb15701.x
- Velázquez I, Pardo JP (2001) Kinetic characterization of the rotenone-insensitive internal NADH: ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Arch Biochem Biophys 389: 7-14. doi:10.1006/abbi.2001.2293