Hyperbolic mixed, dual modification (inhibition → activation)

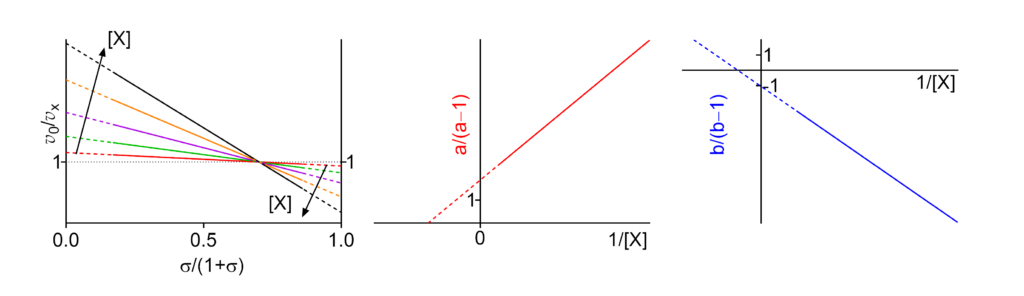
Featured examples
The relative critical substrate concentration σ 0 = (α − β)/(β −1) was calculated for this table from α and β, if provided by the authors.
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors(2) | Reference (3) |
|---|---|---|---|---|---|---|
| 1 | Arachidonate 15-lipoxygenase Homo sapiens | 1.13.11.33 | 13-(S)-HODE | γ-Linolenic acid | K-type inhibition & V-type activation α = 7.3, β = 1.6, KX = 9.8 μM, σ 0 = 9.5 | Joshi (2013) |
| 2 | Arachidonate 15-lipoxygenase Homo sapiens | 1.13.11.33 | 13-(S)-HOTrE(γ) | γ-Linolenic acid | K-type inhibition & V-type activation α = 3.5, β = 1.2, KX= 4.9 μM, σ 0 = 11.5 | Joshi (2013) |
| 3 | Glutathione transferase Mus musculus | 2.5.1.18 | Ethacrinic acid | 1-Chloro-2,4-dinitrobenzene | Anomalous inhibition (4) | Phillips (1991) |
| 4 | Cathepsin K Homo sapiens | 3.4.22.38 | NSC13345 (5) | Abz–HPGGPQ–EDDnp (6) | σ < σ 0 (inhibition), σ > σ 0 (activation) α = 2.4, β = 1.3, KX = 28 μM, σ 0 = 3.7 | Novinec (2014b) |
| 5 | Cathepsin K Homo sapiens | 3.4.22.38 | (2-Biphenylylmethyl) malonic acid | Cbz-Phe-Arg-7-amino-4-methylcoumarylamide | σ < σ 0 (inhibition), σ > σ 0 (activation) α = 5.6, β = 1.5, KX = 100 μM, σ 0 = 8.2 | Novinec (2014a) |
| 6 | Cathepsin K Homo sapiens | 3.4.22.38 | 1,4,7,9B-Tetraazaphenalene | Cbz-Phe-Arg-7-amino-4-methylcoumarylamide | σ < σ 0 (inhibition), σ > σ 0 (activation) α = 4.8, β = 1.5, KX = 20 μM, σ 0 = 6.6 | Novinec (2014a) |
| 7 | Chymotrypsin Not specified | 3.4.21.11 | α-Napthylamine | N-Acetylglycine ethylester | No name (7) | Martinek (1971) |
| 8 | Chymotrypsin Not specified | 3.4.22.11 | Phenol | N-Acetylglycine ethylester | No name (7) | Martinek (1971) |
| 9 | Cholinesterase Homo sapiens | 3.1.1.8 | Benzalkonium | 3-(Acetamido) N,N,N-trimethylanilinium | Inhibition at low [S], activation at high [S] α = 5.7, β = 3.1, KX = 0.05 μM, σ 0 = 1.2 | Masson (2008) |
| 10 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Decamethonium | 7-Acetoxy-4-methylcoumarin | No name (8) KX = 0.87 μM | Berman (1990) |
| 11 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Hexamethonium | 7-Acetoxy-4-methylcoumarin | No name (8) KX = 30 μM | Berman (1990) |
| 12 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Decyltrimethylammonium | 7-Acetoxy-4-methylcoumarin | No name (8) KX = 33 μM | Berman (1990) |
| 13 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Hexyltrimethylammonium | 7-Acetoxy-4-methylcoumarin | No name (8) KX = 200 μM | Berman (1990) |
| 14 | Aldehyde dehydrogenase (NAD+) Homo sapiens | 1.2.1.3 | Alda-1 | Propanal | Inhibition at low [S], activation at high [S] (9) α = 8.6, β = 1.6 | Belmont-Díaz (2016) |
| 15 | Gamma-glutamyl transferase Homo sapiens | 2.3.2.2 | Compound 11 | D-glutamic acid γ-4-nitro- anilide HCl (10) | V-type activation (11) α > β, β = 5.4 | Wickham (2012, 2013) |
| 16 | Gamma-glutamyl transferase Homo sapiens | 2.3.2.2 | Compound 14 | Glutathione (10) | V-type activation (12) β = 4.0 | Wickham (2012, 2013) |
| 17 | Dipeptidyl-peptidase I (tetramer) Homo sapiens | 3.4.14.1 | 3'-Nitrophthalanilic acid | H-Gly-Phe-7-amino-4-methylcoumarylamide | Hyperbolic mixed inhibition/activation α = 3.7, β = 1.9, KX = 130 μM, σ 0 = 2.0 | Rebernik (2019) |
| 18 | beta-Glucosidase Humicola insolens Mutant: D237V/P389H/E395G/K475R | 3.2.1.21 | Glucose | p-nitrophenyl- β-D-glucopyranoside | Stimulation (13) α = 3.3, β = 1.3, Ka = 19.7 mM | Meleiro (2017) |
| 19 | beta-Glucosidase Humicola insolens Mutant: N89Y/H307Y | 3.2.1.21 | Xylose | p-nitrophenyl- β-D-glucopyranoside | Stimulation (13) α = 5.2, β = 3.3, Ka = 31.1 mM | Meleiro (2017) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) ‘…simultaneously as a competitive inhibitor and as an uncompetitive activator’. Fig. 3 (Phillips, 1991) strongly suggest HMxD(I/A) as the mechanism. The authors proposed a mechanism with 12 complexes and 15 parameters.
(5) NSC13345 = 2-[(2-carbamoylsulphanylacetyl)-amino]benzoic acid.
(6) Abz–HPGGPQ–EDDnp = Fluorogenic, internally quenched substrate, where Abz represents o-aminobenzoic acid and EDDnp represents N -(2,4-dinitrophenyl)-ethylenediamine]. Cleavage by cathepsin K occurs at the Gly-Gly bond, Km = 3.8 ± 0.5 μM, kcat = 2.9 ± 0.1 s–1.
(7) The dual behavior of the modifier (called by the authors effector) was demonstrated unambiguously. Parameter determination was not attempted.
(8) In double-reciprocal plots ‘The intersection point occurred in the upper right quadrant’.
(9) ‘Unusual intersection of the lines of the double-reciprocal plot in the first quadrant of the Cartesian plane’.
(10) Hydrolysis reaction of gamma-glutamyltransferase.
(11) The identity of the mechanism, HMxD(I/A), has been unmistakably demonstrated in Figs. 5A and 5B by Wickham (2012). Using the data in Fig. 5A, all dependencies of the kinetic parameters and the specific velocity plot could be drawn. The profiles were identical with those in the ‘Fingerprints’ above. For α only an approximate value could be inferred from the specific velocity plot and this was larger than the value of β calculated by the authors as ‘fold activation’ i.e. as the ratio of the limiting rates at saturating modifier and in the absence of modifier. The reason why only activation, not inhibition, has been observed in the experiment of Figs. 5A and 5B can be sought in the substrate (D-GpNA) concentrations used in the measurements, which spanned from 0.25 to 3.0 mM (Km = 0.16 mM, as reported by the authors), i.e. there were no substrate concentrations below or in the range of Km. Since the critical substrate concentration calculated from the specific velocity plot was in the range 0.03-0.04, the inhibitory part of this system could obliviously not be seen.
(12) Also in this case, the identity of the mechanism, HMxD(I/A), is clear as shown in Fig. 5, Wickham (2013). This is the only one among the 17 basic modifier mechanisms showing hyperbolically increasing Vapp and hyperbolically decreasing Vapp/Kmapp for increasing modifier concentration. A value of α could nor be guessed from Fig. 5 because no raw data have been shown. Note: Fig. 5 shows that Vapp increases in a nearly linear fashion with increasing modifier concentration. As we know, there is no apparent limiting rate (or kcatapp) that can increase linearly with modifier concentration. This is possibly due to high substrate concentrations, close to saturation.
(13) The parameter α was calculated from the Kmapp-values in Table 2, while the values of β are listed in Table 3 as stimulation factor (fold). Ka was reported by the authors as the modifier concentration needed for half-maximal activation.
References
- Belmont-Díaz JA, Yoval-Sánchez B, Calleja-Castañeda LF, Pardo Vázquez JP, Rodríguez-Zavala JS (2016) Alda-1 modulates the kinetic properties of mitochondrial aldehyde dehydrogenase (ALDH2). FEBS J 283: 3637-3650. doi:10.1111/febs.13833
- Berman HA, Leonard K (1990) Ligand exclusion on acetylcholinesterase. Biochemistry 29: 10640-10649. doi:10.1021/bi00499a010
- Joshi N, Hoobler EK, Perry S, Diaz G, Fox B, Holman TR (2013) Kinetic and structural investigations into the allosteric and pH effect on the substrate specificity of human epithelial 15-lipoxygenase-2. Biochemistry 52: 8026-8035. doi:10.1021/bi4010649
- Martinek K, Varfolomeev SD, Levashov AV, Berezin IV (1971) Kinetic manifestations of the structure of the active center of a-chymotrypsin on interacting with fragments of specific substrates. Mol Biol Engl Transl 5: 278-286.
- Masson P, Froment MT, Gillon E, Nachon F, Lockridge O, Schopfer LM (2008) Kinetic analysis of effector modulation of butyrylcholinesterase-catalysed hydrolysis of acetanilides and homologous esters. FEBS J 275: 2617-2631. doi:10.1111/j.1742-4658.2008.06409.x
- Meleiro LP, Salgado JCS, Maldonado RF, Carli S, Moraes LAB, Ward RJ, Jorge JA, Furriel RPM (2017) Engineering the GH1 β-glucosidase from Humicola insolens: Insights on the stimulation of activity by glucose and xylose. PLOS ONE 12: e0188254. doi:10.1371/journal.pone.0188254
- Novinec M, Lenarčič B, Baici A (2014a) Probing the activity modification space of the cysteine peptidase cathepsin K with novel allosteric modifiers. PLoS One 9: e106642. doi:10.1371/journal.pone.0106642
- Novinec M, Korenč M, Caflisch A, Ranganathan R, Lenarčič B, Baici A (2014b) A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods. Nat Commun 5: 4287. doi:10.1038/ncomms4287
- Phillips MF, Mantle TJ (1991) The initial-rate kinetics of mouse glutathione S-transferase YfYf. Evidence for an allosteric site for ethacrynic acid. Biochem J 275: 703-709. doi:10.1042/bj2750703
- Rebernik M, Snoj T, Klemenčič M, Novinec M (2019) Interplay between tetrameric structure, enzymatic activity and allosteric regulation of human dipeptidyl-peptidase I. Arch Biochem Biophys 675: 108121. doi:10.1016/j.abb.2019.108121
- Wickham S, Regan N, West MB, Kumar VP, Thai J, Li PK, Cook PF, Hanigan MH (2012) Divergent effects of compounds on the hydrolysis and transpeptidation reactions of γ-glutamyl transpeptidase. J Enzyme Inhib Med Chem 27: 476-489. doi:10.3109/14756366.2011.597748
- Wickham S, Regan N, West MB, Thai J, Cook PF, Terzyan SS, Li PK, Hanigan MH (2013) Inhibition of human gamma-glutamyl transpeptidase: development of more potent, physiologically relevant, uncompetitive inhibitors. Biochem J 450: 547-557. doi:10.1042/bj20121435