Hyperbolic mixed, predominantly specific activation
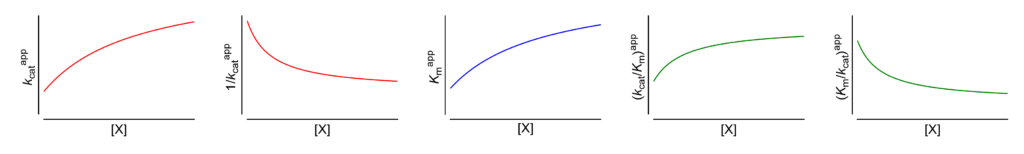
Featured examples
- Km↑ The apparent Michaelis constant increases with increasing [X]
- V ↑ (∴kcat ↑) The apparent limiting rate, and therefore the catalytic constant, increase with increasing [X]
These symbols are shown only when the featured dependencies of the parameters on modifier concentration have been demonstrated by the authors.
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors (2) | Reference(3) |
|---|---|---|---|---|---|---|
| 1 | Phospholipase A2 (Ammodytoxin A) (4) Vipera ammodytes ammodytes | 3.1.1.4 | Calmodulin | PyPG (5) | Nonessential activation α = 4.4, β = 13, KX = 72 pM non-reducing conditions | Kovačič (2009) |
| 2 | Phospholipase A2 (Ammodytoxin A) (4) Vipera ammodytes ammodytes | 3.1.1.4 | Calmodulin | PyPG (5) | Nonessential activation α = 4.4, β = 48, KX = 33 nM reducing (cytosol-like) conditions | Kovačič (2009) |
| 3 | Phospholipase A2 (Ammodytoxin C) (4) Vipera ammodytes ammodytes | 3.1.1.4 | Calmodulin | PyPG (5) | Nonessential activation α = 4.1, β = 6.3, KX = 0.18 nM non-reducing conditions | Kovačič (2009) |
| 4 | Phospholipase A2 (Ammodytoxin C) (4) Vipera ammodytes ammodytes | 3.1.1.4 | Calmodulin | PyPG (5) | Nonessential activation α = 4.1, β = 12, KX = 3.3 nM reducing (cytosol-like) conditions | Kovačič (2009) |
| 5 | 3-Phosphoshikimate 1-carboxyvinyltransferase Bacillus subtilis | 2.5.1.19 | Shikimate-3-phosphate 100 mM NH4Cl | Phosphoenol pyruvate 100 mM NH4Cl | No name K-type inhibition, Km↑ V-type activation, V↑ | Fischer (1987) |
| 6 | 3-Phosphoshikimate 1-carboxyvinyltransferase Bacillus subtilis | 2.5.1.19 | Phosphoenol pyruvate 100 mM NH4Cl | Shikimate-3-phosphate 100 mM NH4Cl | No name K-type inhibition, Km↑ V-type activation, V↑ | Fischer (1987) |
| 7 | Streptopain Streptococcus pyogenes | 3.4.22.10 | Cystatin C (human) (6) | Immunoglobulin G (human) | Activator V↑, Km↑, V/Km↑ | Vincents (2008) |
| 8 | Leukotriene-A4 hydrolase Homo sapiens | 3.3.2.6 | ARM1 | L-Ala-p-nitroanilide | Hyperbolic mixed, predominantly specific activation α = 2.66, β = 23.6, KX = 0.18 μM (7) | Lee (2019) |
| 9 | Coagulation factor VIIa Homo sapiens | 3.4.21.21 | Phosphatidylcholine + phosphatidylserine vesicles | Coagulation factor X Homo sapiens | Activation (8) | Krishnaswamy (1992) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) Ammodytoxin C (less toxic) differs from ammodytoxin A (more toxic) in two amino acid residues
(5) PyPG = 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphoglycerol (large unilamellar vesicles)
(6) An unusual behavior for an expected inhibitor that acts instead as nonessential activator of the target enzyme. Despite few data due to the very difficult system, the dependence of the kinetic parameters on modifier concentration clearly points to HMx(Sp>Ca)A as kinetic mechanism.
(7) The same mechanism with similar kinetic constants is also shared by three other modifiers with structures derived from those of ARM1 (4-(4-benzylphenyl)-thiazol-2-amine).
(8) The authors did not calculate other kinetic parameters besides approximate values of the apparent Michaelis constant and limiting rate at saturating PCPS concentration. There is an error in Figure 8 (the concentration PCPS = 80 μM is plotted at 40 μM, or the value of 80 μM in the figure legend should be 40). Therefore, further calculations were not attempted for this website. However, the dependencies of the kinetic parameters on modifier PCPS concentration clearly point to a predominantly specific activation mechanism, which is assigned to HMx(Sp>Ca)A because the authors reported ‘negligible levels of factor X activation in the absence of PCPS’. Should ‘negligible’ be truly zero, then the mechanism should be assigned to essential activation, LMx(Sp>Ca)A.
References
- Fischer RS, Rubin JL, Gaines CG, Jensen RA (1987) Glyphosate sensitivity of 5-enol-pyruvylshikimate-3-phosphate synthase from Bacillus subtilis depends upon state of activation induced by monovalent cations. Arch Biochem Biophys 256: 325-334. doi:10.1016/0003-9861(87)90453-X
- Lee KH, Petruncio G, Shim A, Burdick M, Zhang Z, Shim YM, Noble SM, Paige M (2019) Effect of modifier structure on the activation of leukotriene A4 hydrolase aminopeptidase activity. J Med Chem 62: 10605-10616. doi:10.1021/acs.jmedchem.9b00663
- Kovačič L, Novinec M, Petan T, Baici A, Križaj I (2009) Calmodulin is a nonessential activator of secretory phospholipase A(2). Biochemistry 48: 11319-11328. doi:10.1021/bi901244f
- Krishnaswamy S, Field KA, Edgington TS, Morrissey JH, Mann KG (1992) Role of the membrane surface in the activation of human coagulation factor X. J Biol Chem 267: 26110-26120.
- Vincents B, Vindebro R, Abrahamson M, von Pawel-Rammingen U (2008) The human protease inhibitor cystatin C is an activating cofactor for the streptococcal cysteine protease IdeS. Chem Biol 15: 960-968. doi:10.1016/j.chembiol.2008.07.021