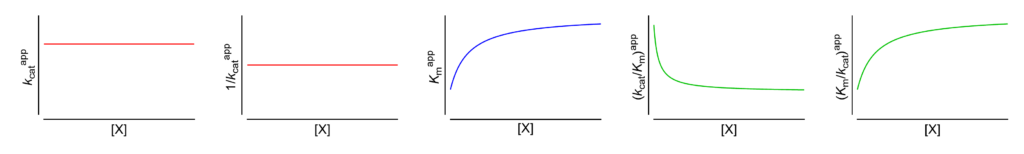
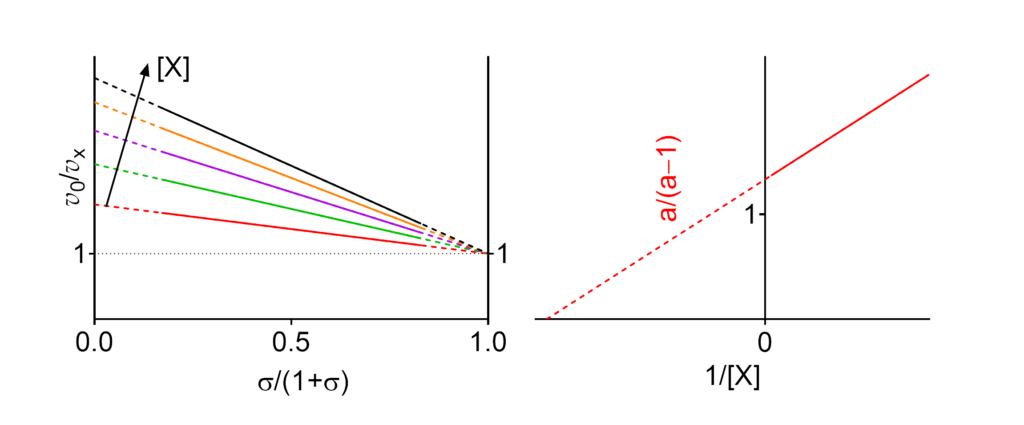
Featured examples
| # | Enzyme Species | EC no. | Modifier | Substrate(1) | Name given by authors (2) | Reference(3) |
|---|---|---|---|---|---|---|
| 1 | Acetylcholinesterase Torpedo californica | 3.1.1.7 | Propidium | 7-acetoxy-4-methyl-coumarin | Nonlinear competitive inhibition α = 6.0, β = 1, KX = 1.2 μM | Berman (1990) |
| 2 | Cathepsin K Homo sapiens | 3.4.22.38 | Compound 6 | Cbz-Phe-Arg-7-amino-4-methyl- coumarylamide | Hyperbolic competitive inhibition α = 3.0, β = 1, KX = 270 μM | Novinec (2014) |
| 3 | Coagulation factor VIIa Homo sapiens | 3.4.21.21 | Peptide A-183 EEWEVLCWTWETCER | N-Methylsulfonyl-D-Phe-Gly-Arg-4-nitroanilide acetate | Partial competitive inhibition α = 2.2, β = 1, KX not calculable | Dennis (2001) |
| 4 | DNA helicase Human papillomavirus 6 E1 | 3.6.4.12 | Bisphenylsulfonaceticacid derivative 4 (4) | ATP | Hyperbolic competitive inhibition α = 70, β = 1, KX = 190 μM | White (2005) |
| 5 | DNA helicase Human papillomavirus 6 E1 | 3.6.4.12 | Bisphenylsulfonaceticacid derivative 6 (4) | ATP | Hyperbolic competitive inhibition α = 11, β = 1, KX = 26 μM | White (2005) |
| 6 | DNA helicase Human papillomavirus 6 E1 | 3.6.4.12 | Bisphenylsulfonaceticacid derivative 7 (4) | ATP | Hyperbolic competitive inhibition α = 10, β = 1, KX = 35 μM | White (2005) |
| 7 | Thrombin Homo sapiens | 3.4.21.5 | Nα-acetyl desulfo hirudin 45-65 | Tos-Gly-Pro-Arg-p-nitroanilide | Partially competitive inhibition α = 4, β = 1, KX = 0.11 μM | DiMaio (1990) |
| 8 | Thrombin Bos taurus | 3.4.21.5 | Nα-acetyl desulfo hirudin 45-65 | Tos-Gly-Pro-Arg-p-nitroanilide | Partially competitive inhibition α = 2, β = 1, KX = 0.72 μM | DiMaio (1990) |
| 9 | NAD( P )H oxidase (5) Mycobacterium tuberculosis | 1.6.3.1 | NADox | NADred | Hyperbolic competitive inhibition α = 6.2±1.7, β = 1, KX = 0.22 mM (6) | Worcel (1965) |
| 10 | Aldehyde dehydrogenase (NAD+) Ovis aries | 1.2.1.3 | Propionaldehyde | 4-Nitrophenylacetate (esterase activity) | Partially competitive inhibition α = 5.3, β = 1, KX = 1.44 μM | Blackwell (1983) |
| 11 | Ferredoxin-nitrite reductase Chlamydomonas reinhardtii (7) | 1.7.7.1 | Nitrate | Nitrite | Partially competitive inhibition α = 12.5, β = 1, KX = 2.7 μM | Córdoba (1986) |
| 12 | Receptor protein-tyrosine kinase Homo sapiens | 2.7.10.1 | RG 14467 | ATP(8) | Hyperbolic competitive inhibition α ≈ 4, β ≈ 1, KX ≤ 30 nM (8) | Hsu (1991) |
| 13 | Leukocyte elastase Homo sapiens | 3.4.21.37 | Heparin (17-19 kDa) | Suc-Ala3-p-nitroanilide | Hyperbolic competitive inhibition α = 3.5, β = 1, KX = 6.8 nM (tight-binding) | Spencer (2006) |
| 14 | Purine nucleosidase Trypanosoma vivax | 3.2.2.2 | Dromedary antibody variable domain fragment (VHH 1602) | p-nitrophenyl riboside | Hyperbolic competitive inhibition α = 2.9, β = 1, KX = 71 nM | Barlow (2009) |
| 15 | Pyruvate kinase Homo sapiens | 2.7.1.40 | L-Phenylalanine | Phosphoenolpyruvate | Hyperbolic specific inhibition α > 1, β ≈ 1, KX = 19 μM | Macpherson (2019) |
(1) Always the varied substrate. In two- or more-substrate reactions the concentration(s) of the non varied substrate(s) is/are kept constant.
(2) Name of the mechanism given by the authors in the quoted reference. α, β and the inhibition/activation constants for the modifier (X), uniformly denoted KX, are the values specified by the authors. In some cases, missing parameters have been calculated from graphical or tabular data provided in the papers. In two- or more-substrate reactions, KX represents an apparent constant at given concentrations of the fixed substrates and no calculations of the intrinsic values have been attempted.
(3) Full references at the end of the page provide also the digital object identifier (doi), if available. Clicking the authors (highlighted) opens the reference in PubMed.
(4) Two experiments are reported in the paper. The values shown here refer to experiment 1.
(5) AMP-activated enzyme.
(6) The mechanism shown in Scheme 2 (p. 3406) lacks the closure of the thermodynamic cycle between E’S and IE’S, necessary for showing the linked functions. α was calculated as K2‘/K1‘, the ratio of the apparent Michaelis constant at saturating [X] and Km in the absence of modifier, from the data below equation 10 on p. 3404.
(7) Wild type, strain 6145c.
(8) At fixed second substrate (K1), a peptide containing the major autophosphorylation site (Tyr-1173) of the epidermal growth factor receptor. Tight-binding (quasi irreversible) slow-onset inhibition; KX estimated by the authors. Approximate values of α and β were estimated from the data in Fig. 10.
References
- Barlow JN, Conrath K, Steyaert J (2009) Substrate-dependent modulation of enzyme activity by allosteric effector antibodies. Biochimica et Biophysica Acta – Proteins and Proteomics 1794: 1259-1268. doi:10.1016/j.bbapap.2009.03.019
- Berman HA, Leonard K (1990) Ligand exclusion on acetylcholinesterase. Biochemistry 29: 10640-10649. doi:10.1021/bi00499a010
- Blackwell LF, Bennett AF, Buckley PD (1983) Relationship between the mechanisms of the esterase and dehydrogenase activities of the cytoplasmic aldehyde dehydrogenase from sheep liver. An alternative view. Biochemistry 22: 3784-3791. doi:10.1021/bi00285a011
- Córdoba F, Cárdenas J, Fernández E (1986) Kinetic characterization of nitrite uptake and reduction by Chlamydomonas reinhardtii. Plant Physiol 82: 904-908. doi:10.1104/pp.82.4.904
- Dennis MS, Roberge M, Quan C, Lazarus RA (2001) Selection and characterization of a new class of peptide exosite inhibitors of coagulation factor VIIa. Biochemistry 40: 9513-9521. doi:10.1021/bi010591l
- DiMaio J, Gibbs B, Munn D, Lefebvre J, Ni F, Konishi Y (1990) Bifunctional thrombin inhibitors based on the sequence of hirudin45-65. J Biol Chem 265: 21698-21703. doi:10.1007/978-94-011-3034-9_321
- Hsu CY, Persons PE, Spada AP, Bednar RA, Levitzki A, Zilberstein A (1991) Kinetic analysis of the inhibition of the epidermal growth factor receptor tyrosine kinase by lavendustin-A and its analogue. J Biol Chem 266: 21105-21112.
- Macpherson JA, Theisen A, Masino L, Fets L, Driscoll PC, Encheva V, Snijders AP, Martin SR, Kleinjung J, Barran PE, Fraternali F, Anastasiou D (2019) Functional cross-talk between allosteric effects of activating and inhibiting ligands underlies PKM2 regulation. eLife 8: e45068. doi:10.7554/eLife.45068
- Novinec M, Lenarčič B, Baici A (2014) Probing the activity modification space of the cysteine peptidase cathepsin K with novel allosteric modifiers. PLoS One 9: e106642. doi:10.1371/journal.pone.0106642
- Spencer JL, Stone PJ, Nugent MA (2006) New insights into the inhibition of human neutrophil elastase by heparin. Biochemistry 45: 9104-9120. doi:10.1021/bi060338r
- White PW, Faucher AM, Massariol MJ, Welchner E, Rancourt J, Cartier M, Archambault J (2005) Biphenylsulfonacetic acid inhibitors of the human papillomavirus type 6 E1 helicase inhibit ATP hydrolysis by an allosteric mechanism involving tyrosine 486. Antimicrobial agents and chemotherapy 49: 4834-4842. doi:10.1128/AAC.49.12.4834-4842.2005
- Worcel A, Goldman DS, Cleland WW (1965) An allosteric reduced nicotinamide adenine dinucleotide oxidase from Mycobacterium tuberculosis. J Biol Chem 240: 3399-3407.